Air sampling for early pathogen detection
Lennart Justen and Will Bradshaw
February 3rd, 2022
This report summarizes an investigation into air sampling as part of a broader effort by the NAO to evaluate different sampling strategies for pathogen early warning.
What is air sampling?
Air sampling is the process of collecting particulate matter from the air with a sampling device. In identifying biological threats, this method is particularly focused on the sampling of airborne microbes, or bioaerosols. The collection and concentration of these bioaerosols enable the analysis of the microbial composition of the sample and its surrounding environment. Techniques such as Polymerase Chain Reaction (PCR) and metagenomic sequencing, similar to those used in wastewater biosurveillance studies, are applied for this purpose. Therefore, air sampling acts as a method of environmental surveillance, targeting materials released by humans into their environment, rather than directly sampling individuals.
Air sampling is particularly promising due to its ability to detect respiratory pathogens, which predominantly spread through the air. This is a key advantage as airborne pathogens are often considered the most likely culprits for future pandemics. Additionally, air samples tend to be less complex compared to other mediums, such as wastewater, and may contain a higher prevalence of human pathogens in relation to other microorganisms. Wastewater, on the other hand, is highly complex and teeming with a vast array of non-pathogenic organisms. These characteristics of air samples potentially make the detection of threats more tractable than in wastewater samples.
In the subsequent sections of this report, we will examine various air sampling methodologies and explore their potential as an effective tool for the early detection of biothreats. Additionally, we will provide a high-level comparison between air sampling and wastewater (WW) sampling, specifically focusing on metagenomic sequencing (MGS)-based pathogen detection.
How are air samples collected?
To collect pathogens and other particulate matter in the air, we need a collection mechanism and a means of moving air over or through it [1]. Air-moving systems (such as fans or vacuums) are typically not very technically challenging and rarely constitute a bottleneck for effective air sampling. Effective collection of particulate matter is decidedly more challenging, with several conflicting performance considerations that must be optimized collectively (Table 1). The ideal collection mechanism for metagenomic sequencing (MGS) surveillance will exhibit high flow rates, high capture and recovery efficiencies, low destructiveness of nucleic acid (NA) in the sample, and low cost. However, tradeoffs between these factors are inevitable [2].
Collection mechanisms can be divided into five major categories (1–3). See also Figure 1 and Appendix 1.
- Filtration: This method employs various physical mechanisms to capture particles of different sizes. The complexity of these mechanisms is considerable and is discussed in greater detail in this report (see page 5).
- Impaction: These methods gather particles that possess sufficient inertia to deviate from an airstream following a sudden directional change.
- Impingement: This technique involves inertial separation into a liquid medium. While liquid mediums generally facilitate easier sample recovery, they can evaporate rapidly under high flow rates.
- Electrostatic Precipitation: This approach charges incoming particles, which are then attracted or repelled towards a collection medium through electrostatic forces.
- Condensation: This method utilizes temperature variations to create water droplets around particles, making them heavy enough to fall into a collection medium.
| Parameter | Description |
|---|---|
| Flow rate | How quickly air can be moved through the device (L/min). Higher flow rates allow faster collection of material and larger/more frequent samples. |
| Collection efficiency | The fraction of particles passing through the sampler that is collected. Typically rated for particular size ranges, e.g., a MERV 13 filter is at least 85% efficient at capturing particles between 1 µm to 3 µm in diameter. |
| Recovery efficiency | The fraction of collected NA that can be recovered from the collection mechanism. |
| Destructiveness (viability) | The degree to which the air sampler desiccates or otherwise kills microbes such that they can no longer be cultured. Critical for culture-based surveillance, less important for sequencing-based surveillance. |
| Destructiveness (fragmentation) | The degree to which an air sampler damages microbial NA through fragmentation or degradation, resulting in the loss of NA in the sample or shorter fragment lengths for sequencing. |
| Cost | Up-front and per-sample costs of buying and using the device. |
| Practicality | Size of sampler, ease of deployment, noise level, etc. |
Table 1: Key performance considerations for air sampling devices
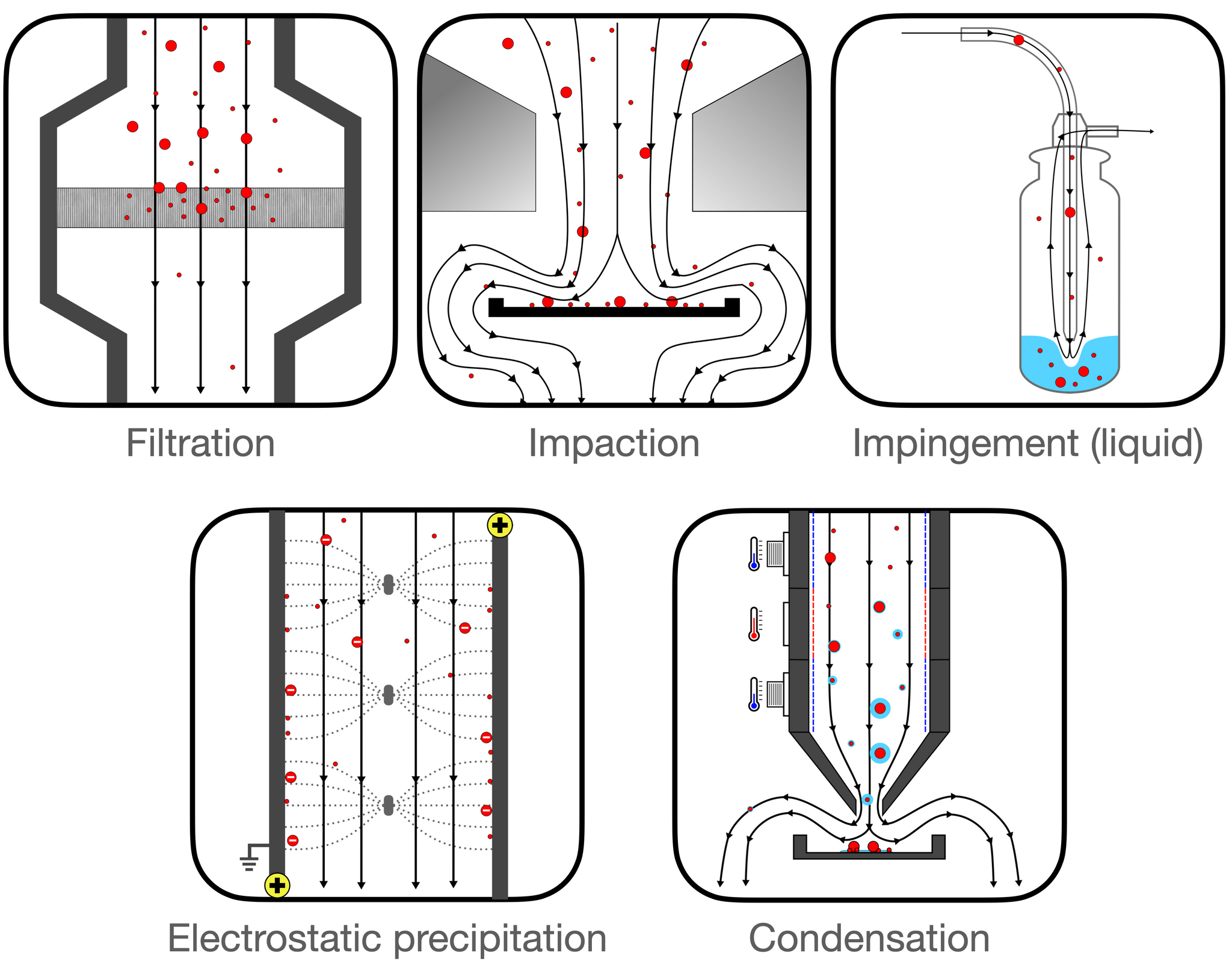
Figure 1: Collection mechanisms employed by different types of air samplers.
Certain air sampling devices incorporate multiple collection mechanisms[3]. The collection mechanism significantly impacts various key performance metrics, as outlined in Table 1, though there is also significant variation within each category.
What is in the air?
The air contains particulate matter from both biological and non-biological sources. Major biological contributors are humans, animals, plants, soil, and mold. Airborne microbes from humans and animals often originate from skin or are expelled as microbe-laden respiratory aerosols and droplets. The size of these particles varies: virus-containing particles (VCPs) are typically 0.02-0.30 µm in diameter, bacteria-containing particles (BCPs) range from 0.5-5 µm, and fungi-containing particles (FCPs) span 1-100 µm in diameter (4).
Air is generally a sparse medium for these particles. Typical indoor concentrations of VCPs and BCPs[4] are about 100 to 1000 particles per liter of air (4), while the count of individual pathogens, as measured by quantitative PCR (qPCR), often falls between 0.1 and 100 copies per liter, as shown in Appendix 2. For context, in January 2023, Biobot reported SARS-CoV-2 levels in Boston's wastewater at approximately 1 million copies per liter, with peaks reaching around 5 million copies per liter during the Omicron surge.
Metagenomic sequencing analysis is a valuable tool for identifying the composition of airborne microbes in specific environments. Generally, the taxonomic complexity in air samples is relatively low, with most studies identifying fewer than 1000 Operational Taxonomic Units (OTUs). In the nine metagenomic studies included in this review, bacteria and human-associated sequences were often the most abundant taxa. It was also common to find a significant proportion (over 50%) of reads or contigs without matches to any known sequences. Viruses were typically found in low relative abundances, usually under 1%, but in some cases, as high as 3.2% (5–10). Within the viral component, bacteriophages were usually the most prevalent, often accounting for over 40% of the viral reads, while human pathogens comprised a smaller percentage. Despite their lower abundance, metagenomic and qPCR studies confirm that human pathogens are detectable in air samples. Identified pathogens in air samples include SARS-CoV-2 and other common coronaviruses (11–21), influenza viruses (21–35), norovirus (36–38), various bacterial pathogens (39–42) including Tuberculosis, and even HIV (6).
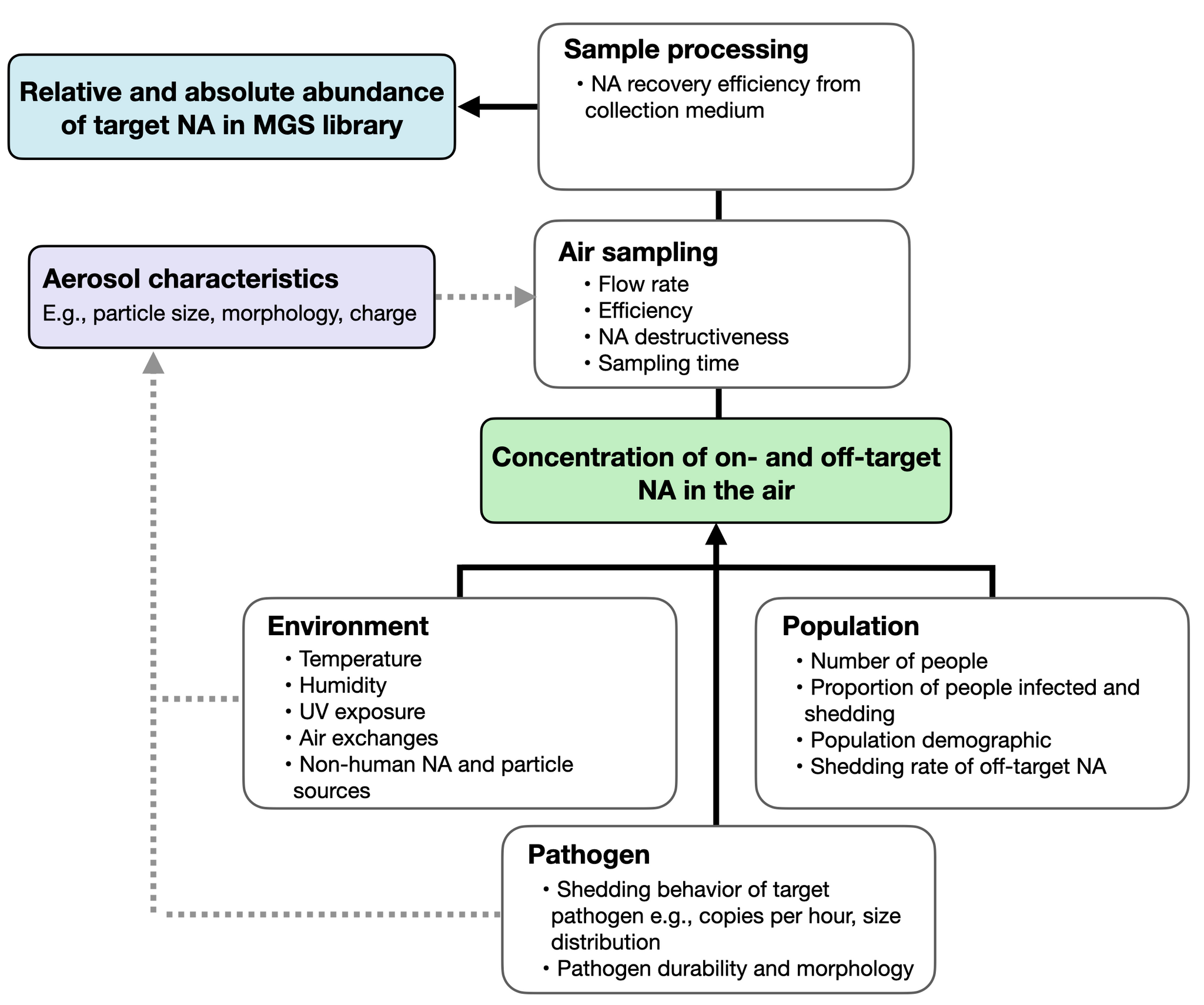
Figure 2: Factors that determine the relative abundance of a target pathogen. Environmental and population parameters are largely determined by choice of location.
What are promising locations for air sampling?
Sampling location is the most crucial factor in determining the success of air sampling for detecting emerging human pathogens. Most obviously, location will determine the physical size of the sampling space and the level of exogenous air movement, as well as environmental conditions like temperature and humidity that influence the viability, movement, and agglomeration behavior of microbes in the air (Figure 2). Location will also determine the size and composition of the human population shedding into the space, significantly affecting infection rates and shedding levels.
Based on these factors, some sampling locations will be far more effective than others for pathogen-agnostic biosurveillance. In particular, indoor and in-duct environments seem significantly more promising than outdoor environments (Appendix 3, 4), as the latter exhibit much more variable environmental conditions and much lower concentrations of human pathogens. High-traffic public buildings like hospitals, airports, and public transit stations are among the most promising locations, as they generally have a large amount of human traffic, favorable sampling demographics, and a relatively straightforward sample collection procedure.
Another promising approach is sampling air from individual airplanes. Airplane cabins contain a high density of people in a small confined space, with a centralized HVAC system (Figure 3) and a high rate of air exchange (13-15 air changes per hour (43)). In most airplanes, cabin air is driven through multiple HEPA filters before being mixed with clean outdoor air and returned to the cabin. While these HEPA filters are usually changed only infrequently (e.g., every 18 months (44)), they are easy to access and replace, and it should be possible to change them more frequently if needed. Taken together, the high density of passengers, unique target population (international travelers), high rates of air exchange, and high capture efficiency of HEPA filters (>99.97% per filter) constitute ideal conditions for collecting high-NA-content air samples.
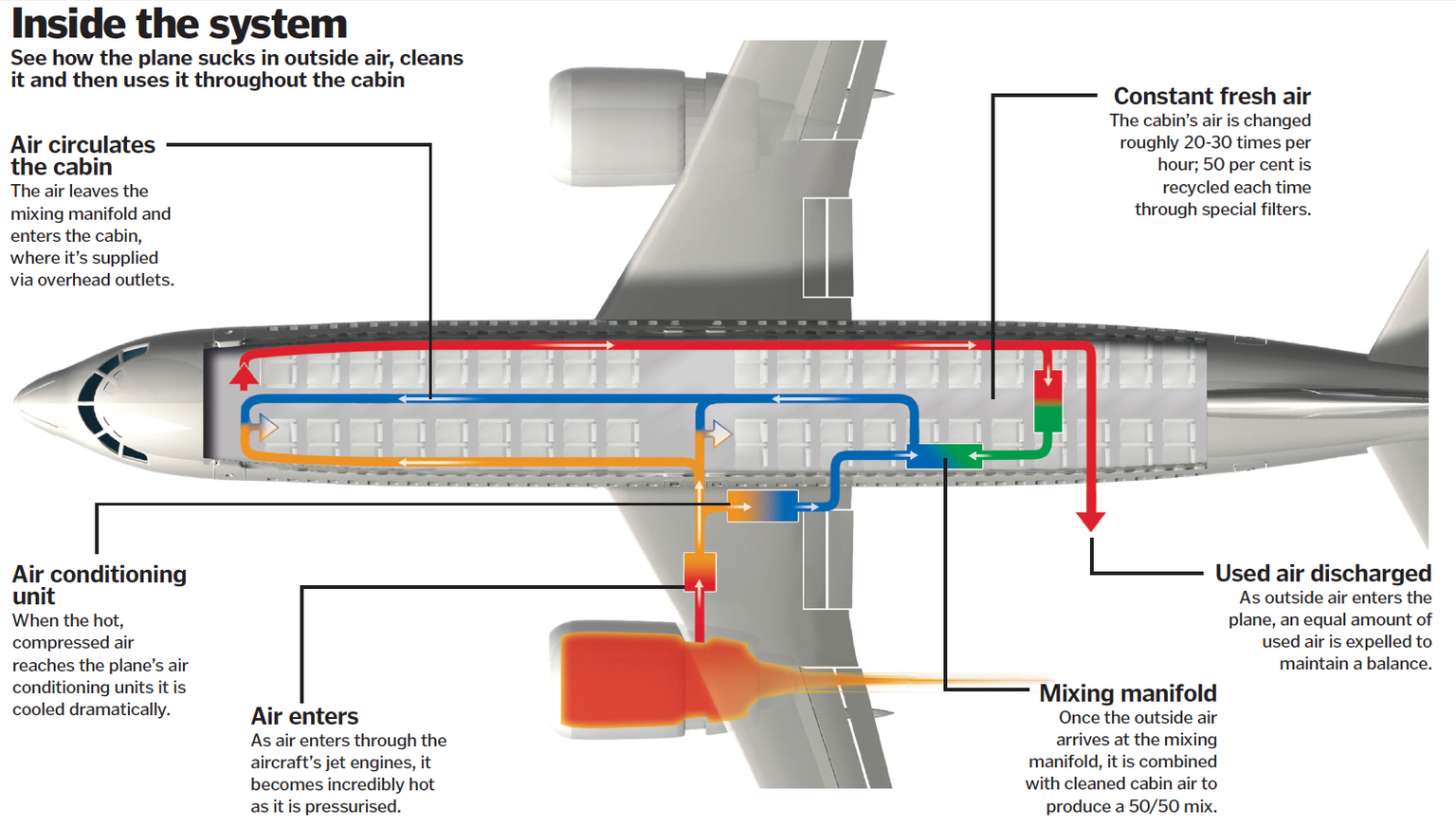
Figure 3: HVAC system aboard an airplane
Conclusion: challenges and opportunities
As a form of environmental surveillance, air sampling could provide a relatively low-effort way to obtain composite samples across many different individuals while maintaining people’s anonymity. Compared to wastewater sampling, air sampling might be better suited to detect pathogens that spread via airborne transmission, both on priors (if a pathogen infects others via airborne transmission, it must be in the air and should therefore be collectible) and as evidenced by numerous studies detecting respiratory pathogens in air samples.
At the same time, air sampling also faces significant challenges. The biggest and most apparent is the sheer sparsity of air as a medium; collecting an adequate amount of airborne NA to perform quality metagenomic sequencing will be a significant challenge[5]. This is exacerbated by the low abundance of viruses in the air microbiome: existing data suggests that viral NA makes up less than 1% of total airborne NA, with human viruses representing an even smaller fraction. Sampling bioaerosols, particularly viral bioaerosols, is complex and often involves significant tradeoffs between factors like rate of air collection, degradation of NA, efficiency at small virus-like particle sizes, and practical implementation considerations.
Table 2 outlines a broader comparison of air sampling compared to wastewater. While air sampling does have some unique advantages, our findings here do not suggest that it is significantly more promising than wastewater for the early detection of unknown outbreaks. Our current sense is that if we are constrained to a single sampling approach, wastewater sampling remains a higher priority than air sampling.
| Axes | Air sampling | Wastewater sampling |
|---|---|---|
| Sample collection | Technically challenging with inherent tradeoffs among desirable sampler qualities. Slow to collect NA. | Trivial at WWTP, though sampling at triturators or individual buildings probably requires custom pipe modifications. Small samples can provide large amounts of NA. |
| Sample complexity | Relatively low number of unique OTUs (typically <1000) in indoor air, and low (<1%) abundance of viral NA from human hosts. | Uncertain, but seems to have more unique OTUs and possibly a lower abundance of viral NA from human hosts. |
| Pathogen coverage | Well suited to detect airborne pathogens which are an important part of the threat space. | Well suited to detect fecal-oral pathogens and also many respiratory pathogens. Probably less robust across respiratory pathogens. |
| Catchment area | Effective surveillance of human pathogens is limited to building-level catchment areas. | Sampling from WWTPs provides city-level pathogen surveillance. |
| Composite traveler samples | Can monitor individual airplanes or airport terminals. | Can collect individual or aggregate airplane samples, as well as airport terminal waste. |
Table 2: Comparison of air and wastewater sampling across several key axis
References
1. Mainelis G. Bioaerosol sampling: Classical approaches, advances, and perspectives. Aerosol Science and Technology. 2020 May 3;54(5):496–519.
2. Lindsley WG, Green BJ, Blachere FM, Martin SB, Law BF, Jensen PA, et al. Sampling and characterization of bioaerosols [Internet]. National Institute for Occupational Safety and Health; 2017 Mar. (Manual of Analytical Methods (NMAM)). Available from: https://www.cdc.gov/niosh/nmam/pdf/chapter-ba.pdf
3. Verreault D, Moineau S, Duchaine C. Methods for Sampling of Airborne Viruses. Microbiology and Molecular Biology Reviews. 2008 Sep;72(3):413–44.
4. Prussin AJ, Marr LC, Bibby KJ. Challenges of studying viral aerosol metagenomics and communities in comparison with bacterial and fungal aerosols. FEMS Microbiol Lett. 2014 Aug;357(1):1–9.
5. Prussin AJ, Torres PJ, Shimashita J, Head SR, Bibby KJ, Kelley ST, et al. Seasonal dynamics of DNA and RNA viral bioaerosol communities in a daycare center. Microbiome. 2019 Apr 1;7(1):53.
6. Be NA, Thissen JB, Fofanov VY, Allen JE, Rojas M, Golovko G, et al. Metagenomic Analysis of the Airborne Environment in Urban Spaces. Microb Ecol. 2015 Feb 1;69(2):346–55.
7. Hall RJ, Leblanc-Maridor M, Wang J, Ren X, Moore NE, Brooks CR, et al. Metagenomic Detection of Viruses in Aerosol Samples from Workers in Animal Slaughterhouses. PLOS ONE. 2013 Aug 14;8(8):e72226.
8. Brisebois E, Veillette M, Dion-Dupont V, Lavoie J, Corbeil J, Culley A, et al. Human viral pathogens are pervasive in wastewater treatment center aerosols. Journal of Environmental Sciences. 2018 May 1;67:45–53.
9. Leung MHY, Tong X, Bøifot KO, Bezdan D, Butler DJ, Danko DC, et al. Characterization of the public transit air microbiome and resistome reveals geographical specificity. Microbiome. 2021 May 26;9(1):112.
10. Cao C, Jiang W, Wang B, Fang J, Lang J, Tian G, et al. Inhalable Microorganisms in Beijing’s PM2.5 and PM10 Pollutants during a Severe Smog Event. Environ Sci Technol. 2014 Feb 4;48(3):1499–507.
11. Ratnesar-Shumate S, Bohannon K, Williams G, Holland B, Krause M, Green B, et al. Comparison of the performance of aerosol sampling devices for measuring infectious SARS-CoV-2 aerosols. Aerosol Science and Technology. 2021 Aug 3;55(8):975–86.
12. Dietz L, Constant DA, Fretz M, Horve PF, Olsen-Martinez A, Stenson J, et al. Exploring Integrated Environmental Viral Surveillance of Indoor Environments: A comparison of surface and bioaerosol environmental sampling in hospital rooms with COVID-19 patients [Internet]. medRxiv; 2021 [cited 2022 Nov 14]. p. 2021.03.26.21254416. Available from: https://www.medrxiv.org/content/10.1101/2021.03.26.21254416v1
13. Kenarkoohi A, Noorimotlagh Z, Falahi S, Amarloei A, Mirzaee SA, Pakzad I, et al. Hospital indoor air quality monitoring for the detection of SARS-CoV-2 (COVID-19) virus. Science of The Total Environment. 2020 Dec 15;748:141324.
14. Angel DM, Gao D, DeLay K, Lin EZ, Eldred J, Arnold W, et al. Development and Application of a Polydimethylsiloxane-Based Passive Air Sampler to Assess Personal Exposure to SARS-CoV-2. Environ Sci Technol Lett. 2022 Feb 8;9(2):153–9.
15. Ramuta MD, Newman CM, Brakefield SF, Stauss MR, Wiseman RW, Kita-Yarbro A, et al. SARS-CoV-2 and other respiratory pathogens are detected in continuous air samples from congregate settings. Nat Commun. 2022 Aug 11;13(1):4717.
16. Sousan S, Fan M, Outlaw K, Williams S, Roper RL. SARS-CoV-2 Detection in air samples from inside heating, ventilation, and air conditioning (HVAC) systems- COVID surveillance in student dorms. American Journal of Infection Control. 2022 Mar 1;50(3):330–5.
17. Lednicky JA, Lauzardo M, Fan ZH, Jutla A, Tilly TB, Gangwar M, et al. Viable SARS-CoV-2 in the air of a hospital room with COVID-19 patients. International Journal of Infectious Diseases. 2020 Nov 1;100:476–82.
18. Horve PF, Dietz L, Northcutt D, Stenson J, Wymelenberg KVD. Evaluation of a bioaerosol sampler for indoor environmental surveillance of Severe Acute Respiratory Syndrome Coronavirus 2. PLOS ONE. 2021 Nov 15;16(11):e0257689.
19. Liu Y, Ning Z, Chen Y, Guo M, Liu Y, Gali NK, et al. Aerodynamic analysis of SARS-CoV-2 in two Wuhan hospitals. Nature. 2020 Jun;582(7813):557–60.
20. Chia PY, Coleman KK, Tan YK, Ong SWX, Gum M, Lau SK, et al. Detection of air and surface contamination by SARS-CoV-2 in hospital rooms of infected patients. Nat Commun. 2020 May 29;11(1):2800.
21. Lednicky JA, Shankar SN, Elbadry MA, Gibson JC, Alam MM, Stephenson CJ, et al. Collection of SARS-CoV-2 Virus from the Air of a Clinic within a University Student Health Care Center and Analyses of the Viral Genomic Sequence. Aerosol Air Qual Res. 2020;20(6):1167–71.
22. Yang W, Elankumaran S, Marr LC. Concentrations and size distributions of airborne influenza A viruses measured indoors at a health centre, a day-care centre and on aeroplanes. J R Soc Interface. 2011 Aug 7;8(61):1176–84.
23. Blachere FM, Lindsley WG, Pearce TA, Anderson SE, Fisher M, Khakoo R, et al. Measurement of airborne influenza virus in a hospital emergency department. Clin Infect Dis. 2009 Feb 15;48(4):438–40.
24. Bui VN, Nguyen TT, Nguyen-Viet H, Bui AN, McCallion KA, Lee HS, et al. Bioaerosol Sampling to Detect Avian Influenza Virus in Hanoi’s Largest Live Poultry Market. Clinical Infectious Diseases. 2019 Mar 5;68(6):972–5.
25. Cao G, Noti JD, Blachere FM, Lindsley WG, Beezhold DH. Development of an improved methodology to detect infectious airborne influenza virus using the NIOSH bioaerosol sampler. J Environ Monit. 2011 Nov 29;13(12):3321–8.
26. Fabian P, McDevitt JJ, DeHaan WH, Fung ROP, Cowling BJ, Chan KH, et al. Influenza Virus in Human Exhaled Breath: An Observational Study. PLOS ONE. 2008 Jul 16;3(7):e2691.
27. Lednicky J, Pan M, Loeb J, Hsieh H, Eiguren-Fernandez A, Hering S, et al. Highly efficient collection of infectious pandemic influenza H1N1 virus (2009) through laminar-flow water based condensation. Aerosol Science and Technology. 2016 Jul 2;50(7):i–iv.
28. Prost K, Kloeze H, Mukhi S, Bozek K, Poljak Z, Mubareka S. Bioaerosol and surface sampling for the surveillance of influenza A virus in swine. Transboundary and Emerging Diseases. 2019;66(3):1210–7.
29. Hermann JR, Hoff SJ, Yoon KJ, Burkhardt AC, Evans RB, Zimmerman JJ. Optimization of a Sampling System for Recovery and Detection of Airborne Porcine Reproductive and Respiratory Syndrome Virus and Swine Influenza Virus. Applied and Environmental Microbiology. 2006 Jul;72(7):4811–8.
30. Li J, Leavey A, Wang Y, O’Neil C, Wallace MA, Burnham CAD, et al. Comparing the performance of 3 bioaerosol samplers for influenza virus. Journal of Aerosol Science. 2018 Jan 1;115:133–45.
31. Lindsley WG, Blachere FM, Davis KA, Pearce TA, Fisher MA, Khakoo R, et al. Distribution of Airborne Influenza Virus and Respiratory Syncytial Virus in an Urgent Care Medical Clinic. Clinical Infectious Diseases. 2010 Mar 1;50(5):693–8.
32. Lindsley WG, Blachere FM, Thewlis RE, Vishnu A, Davis KA, Cao G, et al. Measurements of Airborne Influenza Virus in Aerosol Particles from Human Coughs. PLOS ONE. 2010 Nov 30;5(11):e15100.
33. Milton PF JJ McDevitt, EA Houseman, DK. Airborne influenza virus detection with four aerosol samplers using molecular and infectivity assays: considerations for a new infectious virus aerosol sampler. Indoor Air. 2009 Oct 1;19(5):433–41.
34. Pan M, Bonny TS, Loeb J, Jiang X, Lednicky JA, Eiguren-Fernandez A, et al. Collection of Viable Aerosolized Influenza Virus and Other Respiratory Viruses in a Student Health Care Center through Water-Based Condensation Growth. mSphere. 2017 Oct 11;2(5):e00251-17.
35. Wu Y, Shen F, Yao M. Use of gelatin filter and BioSampler in detecting airborne H5N1 nucleotides, bacteria and allergens. Journal of Aerosol Science. 2010 Sep 1;41(9):869–79.
36. Boles C, Brown G, Nonnenmann M. Determination of murine norovirus aerosol concentration during toilet flushing. Sci Rep. 2021 Dec 7;11(1):23558.
37. Bonifait L, Charlebois R, Vimont A, Turgeon N, Veillette M, Longtin Y, et al. Detection and Quantification of Airborne Norovirus During Outbreaks in Healthcare Facilities. Clinical Infectious Diseases. 2015 Aug 1;61(3):299–304.
38. Uhrbrand K, Schultz AC, Madsen AM. Exposure to Airborne Noroviruses and Other Bioaerosol Components at a Wastewater Treatment Plant in Denmark. Food Environ Virol. 2011 Dec 1;3(3):130–7.
39. Gast RK, Mitchell BW, Holt PS. Detection of Airborne Salmonella enteritidis in the Environment of Experimentally Infected Laying Hens by an Electrostatic Sampling Device. avdi. 2004 Jan;48(1):148–54.
40. Wan GH, Huang CG, Huang YC, Huang JP, Yang SL, Lin TY, et al. Surveillance of Airborne Adenovirus and Mycoplasma pneumoniae in a Hospital Pediatric Department. PLOS ONE. 2012 Mar 21;7(3):e33974.
41. Mastorides SM, Oehler RL, Greene JN, Sinnott JT, Kranik M, Sandin RL. The Detection of Airborne Mycobacterium tuberculosis Using Micropore Membrane Air Sampling and Polymerase Chain Reaction. CHEST. 1999 Jan 1;115(1):19–25.
42. Van Droogenbroeck C, Van Risseghem M, Braeckman L, Vanrompay D. Evaluation of bioaerosol sampling techniques for the detection of Chlamydophila psittaci in contaminated air. Veterinary Microbiology. 2009 Mar 16;135(1):31–7.
43. Stuart Walkinshaw D. A Brief Introduction To Passenger Aircraft Cabin Air Quality. ASHRAE Journal. 2020 Oct;12–8.
44. Conrad D. HEPA Filters: Keeping Cabin AIr Safe for Airlines [Internet]. PTI Technologies; 2020 Jun [cited 2023 Jan 31]. Available from: https://www.ptitechnologies.com/wp-content/uploads/2020/12/HEPA-Filters-Keeping-Cabin-Air-Safe-For-Airlines-13-July-2020-reformatted.pdf
45. Lindsley WG. Filter Pore Size and Aerosol Sample Collection. National Institute for Occupational Safety and Health; 2016 Apr. (Manual of Analytical Methods (NMAM)). Report No.: 5th Edition.
46. Burton NC, Grinshpun SA, Reponen T. Physical Collection Efficiency of Filter Materials for Bacteria and Viruses. The Annals of Occupational Hygiene. 2007 Mar 1;51(2):143–51.
47. Mbareche H, Veillette M, Bilodeau GJ, Duchaine C. Bioaerosol Sampler Choice Should Consider Efficiency and Ability of Samplers To Cover Microbial Diversity. Applied and Environmental Microbiology. 2018 Nov 15;84(23):e01589-18.
48. Tseng CC, Li CS. Collection efficiencies of aerosol samplers for virus-containing aerosols. Journal of Aerosol Science. 2005 May 1;36(5):593–607.
49. Verreault D, Rousseau GM, Gendron L, Massé D, Moineau S, Duchaine C. Comparison of Polycarbonate and Polytetrafluoroethylene Filters for Sampling of Airborne Bacteriophages. Aerosol Science and Technology. 2010 Feb 10;44(3):197–201.
50. Gendron L, Verreault D, Veillette M, Moineau S, Duchaine C. Evaluation of Filters for the Sampling and Quantification of RNA Phage Aerosols. Aerosol Science and Technology. 2010 Aug 9;44(10):893–901.
51. Borges JT, Nakada LYK, Maniero MG, Guimarães JR. SARS-CoV-2: a systematic review of indoor air sampling for virus detection. Environ Sci Pollut Res Int. 2021;28(30):40460–73.
52. Choi DY, Heo KJ, Kang J, An EJ, Jung SH, Lee BU, et al. Washable antimicrobial polyester/aluminum air filter with a high capture efficiency and low pressure drop. Journal of Hazardous Materials. 2018 Jun 5;351:29–37.
53. Zhen H, Han T, Fennell DE, Mainelis G. Release of Free DNA by Membrane-Impaired Bacterial Aerosols Due to Aerosolization and Air Sampling. Applied and Environmental Microbiology. 2013 Dec 15;79(24):7780–9.
54. Mainelis G, Tabayoyong M. The Effect of Sampling Time on the Overall Performance of Portable Microbial Impactors. Aerosol Science and Technology. 2010 Jan 1;44(1):75–82.
55. Wang Z, Reponen T, A. Grinshpun S, L. Górny R, Willeke K. Effect of sampling time and air humidity on the bioefficiency of filter samplers for bioaerosol collection. Journal of Aerosol Science. 2001 May 1;32(5):661–74.
56. Hinds WC, Zhu Y. Aerosol Technology: Properties, Behavior, and Measurement of Airborne Particles. John Wiley & Sons; 2022. 452 p.
57. Gerone PJ, Couch RB, Keefer GV, Douglas RG, Derrenbacher EB, Knight V. Assessment of experimental and natural viral aerosols. Bacteriological Reviews. 1966 Sep;30(3):576–88.
58. Stewart SL, Grinshpun SA, Willeke K, Terzieva S, Ulevicius V, Donnelly J. Effect of impact stress on microbial recovery on an agar surface. Applied and Environmental Microbiology. 1995 Apr;61(4):1232–9.
59. Harstad JB. Sampling Submicron T1 Bacteriophage Aerosols. Applied Microbiology. 1965 Nov;13(6):899–908.
60. Lin X, Reponen T, Willeke K, Wang Z, Grinshpun SA, Trunov M. Survival of Airborne Microorganisms During Swirling Aerosol Collection. Aerosol Science and Technology. 2000 Mar;32(3):184–96.
61. Lemieux J, Veillette M, Mbareche H, Duchaine C. Re-aerosolization in liquid-based air samplers induces bias in bacterial diversity. Aerosol Science and Technology. 2019 Nov 2;53(11):1244–60.
62. Tseng CC, Hsiao PK, Chang KC, Chen WT, Yiin LM, Hsieh CJ. Optimization of Propidium Monoazide Quantitative PCR for Evaluating Performances of Bioaerosol Samplers for Sampling Airborne Staphylococcus aureus. Aerosol Science and Technology. 2014 Dec 2;48(12):1308–19.
63. McFarland AR, Haglund JS, King MD, Hu S, Phull MS, Moncla BW, et al. Wetted Wall Cyclones for Bioaerosol Sampling. Aerosol Science and Technology. 2010 Feb 25;44(4):241–52.
64. Berry CM. An Electrostatic Method for Collecting Bacteria from Air. Public Health Reports (1896-1970). 1941;56(42):2044–51.
65. Degois J, Dubuis ME, Turgeon N, Veillette M, Duchaine C. Condensation sampler efficiency for the recovery and infectivity preservation of viral bioaerosols. Aerosol Science and Technology. 2021 Jun 3;55(6):653–64.
66. Hering SV, Stolzenburg MR. A Method for Particle Size Amplification by Water Condensation in a Laminar, Thermally Diffusive Flow. Aerosol Science and Technology. 2005 May 1;39(5):428–36.
67. Hering SV, Spielman SR, Lewis GS. Moderated, Water-Based, Condensational Particle Growth in a Laminar Flow. Aerosol Science and Technology. 2014 Apr 3;48(4):401–8.
68. Lednicky J, Pan M, Loeb J, Hsieh H, Eiguren-Fernandez A, Hering S, et al. Highly efficient collection of infectious pandemic influenza H1N1 virus (2009) through laminar-flow water based condensation. Aerosol Science and Technology. 2016 Jul 2;50(7):i–iv.
69. Nieto-Caballero M, Savage N, Keady P, Hernandez M. High fidelity recovery of airborne microbial genetic materials by direct condensation capture into genomic preservatives. Journal of Microbiological Methods. 2019 Feb 1;157:1–3.
70. Pan M, Carol L, Lednicky JA, Eiguren-Fernandez A, Hering S, Fan ZH, et al. Determination of the distribution of infectious viruses in aerosol particles using water-based condensational growth technology and a bacteriophage MS2 model. Aerosol Science and Technology. 2019 May 4;53(5):583–93.
71. Pan M, Eiguren-Fernandez A, Hsieh H, Afshar-Mohajer N, Hering S v., Lednicky J, et al. Efficient collection of viable virus aerosol through laminar-flow, water-based condensational particle growth. Journal of Applied Microbiology. 2016;120(3):805–15.
72. Eiguren-Fernandez A, Lewis GS, Spielman SR, Hering SV. Time-resolved characterization of particle associated polycyclic aromatic hydrocarbons using a newly-developed sequential spot sampler with automated extraction and analysis. Atmospheric Environment. 2014 Oct 1;96:125–34.
73. Guo J, Xiong Y, Kang T, Xiang Z, Qin C. Bacterial community analysis of floor dust and HEPA filters in air purifiers used in office rooms in ILAS, Beijing. Sci Rep. 2020 Apr 14;10(1):6417.
74. King P, Pham LK, Waltz S, Sphar D, Yamamoto RT, Conrad D, et al. Longitudinal Metagenomic Analysis of Hospital Air Identifies Clinically Relevant Microbes. PLOS ONE. 2016 Aug 2;11(8):e0160124.
75. Meadow JF, Altrichter AE, Kembel SW, Kline J, Mhuireach G, Moriyama M, et al. Indoor airborne bacterial communities are influenced by ventilation, occupancy, and outdoor air source. Indoor Air. 2014;24(1):41–8.
76. Whon TW, Kim MS, Roh SW, Shin NR, Lee HW, Bae JW. Metagenomic Characterization of Airborne Viral DNA Diversity in the Near-Surface Atmosphere. J Virol. 2012 Aug;86(15):8221–31.
77. Rosario K, Fierer N, Miller S, Luongo J, Breitbart M. Diversity of DNA and RNA Viruses in Indoor Air As Assessed via Metagenomic Sequencing. Environ Sci Technol. 2018 Feb 6;52(3):1014–27.
78. Prussin AJ, Garcia EB, Marr LC. Total Virus and Bacteria Concentrations in Indoor and Outdoor Air. Environ Sci Technol Lett. 2015;2(4):84–8.
79. Fang Z, Ouyang Z, Zheng H, Wang X. Concentration and Size Distribution of Culturable Airborne Microorganisms in Outdoor Environments in Beijing, China. Aerosol Science and Technology. 2008 Mar 31;42(5):325–34.
80. Wang S, Liu W, Li J, Sun H, Qian Y, Ding L, et al. Seasonal Variation Characteristics of Bacteria and Fungi in PM2.5 in Typical Basin Cities of Xi’an and Linfen, China. Atmosphere. 2021 Jul;12(7):809.
81. Ho HM, Rao CY, Hsu HH, Chiu YH, Liu CM, Chao HJ. Characteristics and determinants of ambient fungal spores in Hualien, Taiwan. Atmospheric Environment. 2005 Oct 1;39(32):5839–50.
82. Fahlgren C, Hagström Å, Nilsson D, Zweifel UL. Annual Variations in the Diversity, Viability, and Origin of Airborne Bacteria. Appl Environ Microbiol. 2010 May;76(9):3015–25.
83. Tanaka D, Terada Y, Nakashima T, Sakatoku A, Nakamura S. Seasonal variations in airborne bacterial community structures at a suburban site of central Japan over a 1-year time period using PCR-DGGE method. Aerobiologia. 2015 Jun 1;31(2):143–57.
84. Yan D, Zhang T, Su J, Zhao LL, Wang H, Fang XM, et al. Diversity and Composition of Airborne Fungal Community Associated with Particulate Matters in Beijing during Haze and Non-haze Days. Frontiers in Microbiology [Internet]. 2016 [cited 2022 Dec 6];7. Available from: https://www.frontiersin.org/articles/10.3389/fmicb.2016.00487
85. Goudarzi G, Shirmardi M, Khodarahmi F, Hashemi-Shahraki A, Alavi N, Ankali KA, et al. Particulate matter and bacteria characteristics of the Middle East Dust (MED) storms over Ahvaz, Iran. Aerobiologia. 2014 Dec 1;30(4):345–56.
86. Fang Z, Yao W, Lou X, Hao C, Gong C, Ouyang Z. Profile and Characteristics of Culturable Airborne Bacteria in Hangzhou, Southeast of China. Aerosol Air Qual Res. 2016;16(7):1690–700.
87. Serrano-Silva N, Calderón-Ezquerro MC. Metagenomic survey of bacterial diversity in the atmosphere of Mexico City using different sampling methods. Environmental Pollution. 2018 Apr 1;235:20–9.
88. Emory A, Light F. EPA Needs to Fulfill Its Designated Responsibilities to Ensure Effective BioWatch Program [Internet]. 2005 Mar p. 27. Report No.: 2005-P-00012. Available from: https://www.epa.gov/sites/default/files/2015-12/documents/20050323-2005-p-00012.pdf
89. United States Government Accountability Office. Biodefense: DHS Exploring New Methods to Replace BioWatch and Could Benefit from Additional Guidance. 2021 May. Report No.: GAO-21-292.
90. Bonetta Sa, Bonetta Si, Mosso S, Sampò S, Carraro E. Assessment of microbiological indoor air quality in an Italian office building equipped with an HVAC system. Environ Monit Assess. 2010 Feb 1;161(1):473–83.
91. Farnsworth JE, Goyal SM, Kim SW, Kuehn TH, Raynor PC, Ramakrishnan MA, et al. Development of a method for bacteria and virus recovery from heating, ventilation, and air conditioning (HVAC) filters. J Environ Monit. 2006 Oct 5;8(10):1006–13.
92. Maestre JP, Jennings W, Wylie D, Horner SD, Siegel J, Kinney KA. Filter forensics: microbiota recovery from residential HVAC filters. Microbiome. 2018 Jan 30;6(1):22.
93. Stanley NJ, Kuehn TH, Kim SW, Raynor PC, Anantharaman S, Ramakrishnan MA, et al. Background culturable bacteria aerosol in two large public buildings using HVAC filters as long term, passive, high-volume air samplers. J Environ Monit. 2008 Apr;10(4):474–81.
94. Hoisington AJ, Maestre JP, King MD, Siegel JA, Kinney KA. Impact of sampler selection on the characterization of the indoor microbiome via high-throughput sequencing. Building and Environment. 2014 Oct 1;80:274–82.
95. Noris F, Siegel JA, Kinney KA. Evaluation of HVAC filters as a sampling mechanism for indoor microbial communities. Atmospheric Environment. 2011 Jan 1;45(2):338–46.
96. Goyal SM, Anantharaman S, Ramakrishnan MA, Sajja S, Kim SW, Stanley NJ, et al. Detection of viruses in used ventilation filters from two large public buildings. American Journal of Infection Control. 2011 Sep 1;39(7):e30–8.
97. Leung MHY, Wilkins D, Li EKT, Kong FKF, Lee PKH. Indoor-Air Microbiome in an Urban Subway Network: Diversity and Dynamics. Appl Environ Microbiol. 2014 Nov;80(21):6760–70.
Appendix 1: Air sampling methods
Filtration and filter-based samplers
Filter-based bioaerosol collection is one of the most common methods in the field of bioaerosol sampling due to its ease of use, sensitivity to small particle sizes, and ability to process arbitrarily large volumes of air. In this approach, an air mover drives air through a filter that collects particulate matter via several physical mechanisms including interception, electrostatic attraction, and diffusion, all of which are explored in more depth here (pg. 5) (45). The air mover and filter may be combined inside a single device, or the two may be separate, like an HVAC filter placed in front of a vent. The advantage of this separability is that it allows you can collect an essentially arbitrary amount of air with a filter-based approach as long as the air-moving system is capable of providing the throughput.
While the concept of filter-based bioaerosol sampling is relatively simple, collection devices vary widely, and the exact choice of device will depend on the particles being targeted (e.g., smog, viruses, bacteria, etc.) and the downstream analysis being performed. In particular, the choice of filter material, thickness, and porosity all influence a) the recovery efficiency of various particle sizes and morphologies, b) NA fragmentation and microbe viability, and c) the sampler’s power usage[6] (30,45–50). Many different types of filters that have been used for bioaerosol collection include polycarbonate, mixed cellulose ester, polytetrafluoroethylene (Teflon, PTFE), polyvinyl chloride (PVC), nylon, gelatin, and others. To the best of my knowledge, there is no authoritative source on which type of filter is best suited for viral bioaerosol collection, although comparisons of different kinds of filters for viral bioaerosol recovery do exist (11,30,42,46,51). In a dated but well-regarded review of different methods for sampling airborne viruses, (3) suggested that 0.3-μm PTFE filters may be the best option for sampling 10- to 900-nm-diameter virus-laden particles.
Pros
-
Filter-based air samplers are well suited to process large volumes of air, which allows for more NA collection and sampling at greater frequency.
-
According to (3), filters appear to provide the best physical recovery of nanoscale-size particles, which include many viral aerosols. Even though filters often have effective pore sizes greater than nanoscale diameters, it’s diffusion, a Brownian motion phenomenon, rather than interception, where particles larger than the pore size get blocked, that most often facilitates collection of nanoscale particles on filters (45).
Cons
-
A key challenge with many filter-based methods is NA recovery. Simple filter elution techniques often have low NA recovery efficiency, while more aggressive extraction techniques like bead beating can over-fragment NA. I suspect that if you sample enough volume, the NA recovery challenges with filters are not prohibitive and that approaches like dissolvable or washable (52) filters can circumvent some of these challenges. Other approaches to NA recovery from filters include vortexing and ultrasonic agitation.
-
Filter-based samplers (with the exception of gelatin filters) are poor at maintaining microbe viability and are therefore not well suited for culture-based analysis (1,3). It has been suggested that filter-based approaches are more likely to cause structural damage like membrane degradation (53) and increase the virus desiccation, leading to infectivity loss (3). It has also been shown that prolonged sample collection time increases the loss of microbe viability (54,55). Fortunately, for metagenomics, desiccation and infectivity loss are not issues as long as the virus genomes remain intact.
Impaction-based samplers
Another widely used method for collecting bioaerosols is impaction-based sampling, which utilizes the inertia of particles to separate them from air (1). In this technique, air is drawn into the sampler through narrow openings, directed toward a solid surface, and then abruptly changes direction. Particles with sufficient inertia deviate from the airflow and adhere to the solid collection medium[7]. Common mediums include filters, agar plates for direct culturing, and glass slides or other solid surfaces for microscopy.
Impaction-based samplers offer distinctive advantages in studying air microbiomes. For instance, linking multiple impaction-based samplers together forms a “multi-stage impactor.” Each stage in this setup captures progressively smaller particles, enabling size-based separation of airborne particles. This arrangement facilitates enrichment of smaller, virus-like particles and allows for assessing microbial concentration and composition at various particle sizes (3, 48, 57). Another notable feature is the ability of these samplers to measure bioaerosol concentrations over time by capturing particles on a rotating medium, typically an agar plate (3).
While impaction-based samplers are beneficial for culture-based analysis, their distinction from filter-based samplers becomes less clear when used to collect bioaerosols for sequencing via a filter. Integrating filters for NA collection with inertia-based particle separation may enrich virus particle samples, but it can also increase the complexity of the device and impose limitations on air flow rates.
Pros
-
Effective separation and identification of different particle sizes, enhancing the potential for virus-laden particle enrichment.
-
Suitable for culture-based analysis, allowing airborne particles to impact on agar plates that can be directly transferred to an incubator. However, this is not applicable for MGS sample collection.
-
Capability to track aerosol concentrations over time using a rotating collection medium, typically an agar plate.
Cons
- The abrupt deceleration during impaction can cause physical damage to microbes (53, 55, 58).
- Using filters for NA collection in conjunction with inertia-based separation can complicate the sampler and reduce air intake rates.
Liquid-based samplers
Liquid-based samplers, often referred to as liquid impingers, use an inertial mechanism similar to impaction-based samplers for bioaerosol collection. The key difference lies in the collection medium, which is liquid rather than solid. The primary advantage of liquid mediums is the facilitation of simpler sample processing techniques and more efficient recovery of collected microbes and nucleic acids (NA). Additionally, liquid mediums help prevent microbe desiccation, which is advantageous for downstream analyses that require maintaining the infectivity of recovered particles (3, 59).
Nonetheless, common liquid-based samplers have notable drawbacks. Without periodic refilling, liquid mediums are prone to evaporation, limiting the duration of collection or necessitating reduced air intake rates to mitigate rapid evaporation. Furthermore, the evaporation of the liquid medium and the high impaction velocity of airborne particles can lead to particle re-aerosolization or damage upon impact (60, 61).
To address concerns about microbe damage and re-aerosolization, wetted-wall cyclone samplers have been developed. These devices incorporate impingement with centrifugal motion. Here, collection nozzles are angled above the liquid, creating a vortex motion during sampling that coats the container walls with a thin liquid film. Particles impact tangentially with the swirling liquid, reducing impact stress (62). Cyclone samplers are capable of higher flow rates (e.g., 1250 L/min, as noted in reference 63) compared to other liquid-based samplers and can operate for several hours, with the potential for extended duration through built-in liquid level control (1).
Pros
-
Liquid mediums allow for easier sample preparation and enhanced NA recovery efficiency.
-
Least destructive collection method, preventing desiccation and improving recovery of infectious viral particles (3, 59).
-
Can be used in a multi-stage format to separate and identify particle sizes.
Cons
- High impaction velocities and liquid evaporation may compromise microbe viability for infectivity assays and cause particle re-aerosolization (60, 61).
- Some critiques point to the typically low flow rates of liquid-based samplers, although this does not take into account the capabilities of cyclone-class collectors.
Electrostatic-based samplers
Electrostatic collectors, also known as Electrostatic Precipitators (ESPs), capture airborne particles by first drawing them into the device, then imparting an electrostatic charge, and finally depositing them onto a collection medium through electrostatic attraction or repulsion (1). ESPs are versatile, compatible with various deposition mediums such as agar, liquid, and solid surfaces, which allows for the customization of the collection medium based on the requirements of downstream analysis.
ESPs offer several beneficial properties for bioaerosol sampling. Firstly, they deposit particles at significantly lower velocities than inertia-based methods, reducing the likelihood of microbe damage (1, 53). Secondly, ESPs do not require a substantial pressure drop for particle collection, as they are "open channel" devices[8]. This results in lower power consumption while still enabling the collection of large volumes of air. An ESP developed by the U.S. Army in the 1960s, for instance, achieved an impressive flow rate of 10,000 liters per minute (57). Despite these advantages, ESPs have seen limited investigation and commercialization compared to solid and liquid-based impaction or filter-based methods. Mainelis (2019) suggests that this may be due to the rapid advancements in other bioaerosol collection techniques (1).
A primary concern with ESPs, aside from their limited exploration and commercial availability, is their tendency to generate ozone during particle charging. The extent of this issue is unclear, but it has been raised that the ozone produced might compromise microbe integrity and viability, and in certain environments, could even pose a risk to human health due to elevated ozone levels (1, 64).
Pros
-
Gentle on collected samples compared to other methods.
-
Compatible with a variety of collection mediums, allowing for customization.
-
Requires less power for operation.
-
Capable of achieving high flow rates.
Cons
- Relatively under-investigated, possibly due to advancements in alternative collection methods.
- Generation of ozone during particle charging, which may degrade microbes and pose health risks in poorly ventilated areas (64).
- Limited availability of ESPs on the commercial market.
Condensation samplers
Condensation samplers employ a phase change from gas to liquid to capture airborne particles (34, 65–71). Fundamentally, condensation sampling involves moving air through a growth tube with varied temperature zones. This process leads to the formation of water droplets around particulate matter, which then fall onto a liquid or solid collection medium.
A notable example of condensation-based sampling is the Spot Sampler by Aerosol Devices. This device uses a three-stage growth tube for particle capture, described as follows:
-
Conditioner Stage: Air first enters the conditioner, a cool (5°C) wet wall section of the growth tube, where incoming particles acquire a thin water film.
-
Initiator Stage: The air then progresses to the initiator, where additional water vapor is introduced. Here, higher temperatures (35°C) create a supersaturated environment, encouraging larger droplet formation around particles.
-
Moderator Stage: In the moderator at 12°C, excess water vapor is removed, permitting continued droplet growth.
-
Exit: Air then exits the growth tube, with droplets impacting onto a collection medium.
Pros
-
Condensation samplers are highly efficient at collecting small particles. One study reported over 90% efficiency in collecting 5 nm particles (72), while another indicated the potential to capture particles as small as 2 nm (66). This implies that condensation sampling could effectively collect the smallest airborne viral particles.
-
Unlike traditional liquid- and filter-based methods, condensation sampling avoids high-velocity impacts, making it a non-destructive technique ideal for preserving microbe viability and genomic integrity.
Cons
-
Condensation samplers typically have low flow rates. For instance, the Spot Sampler operates at only 1.5 L/min, posing a challenge in collecting sufficient NA samples in short timeframes.
-
The market and research base for condensation samplers seem relatively underdeveloped, as indicated by their limited discussion in two comprehensive review papers on bioaerosol sampling (1, 3).
Appendix 2: Microbe concentrations
See also Zhai et al. 2018 for review.
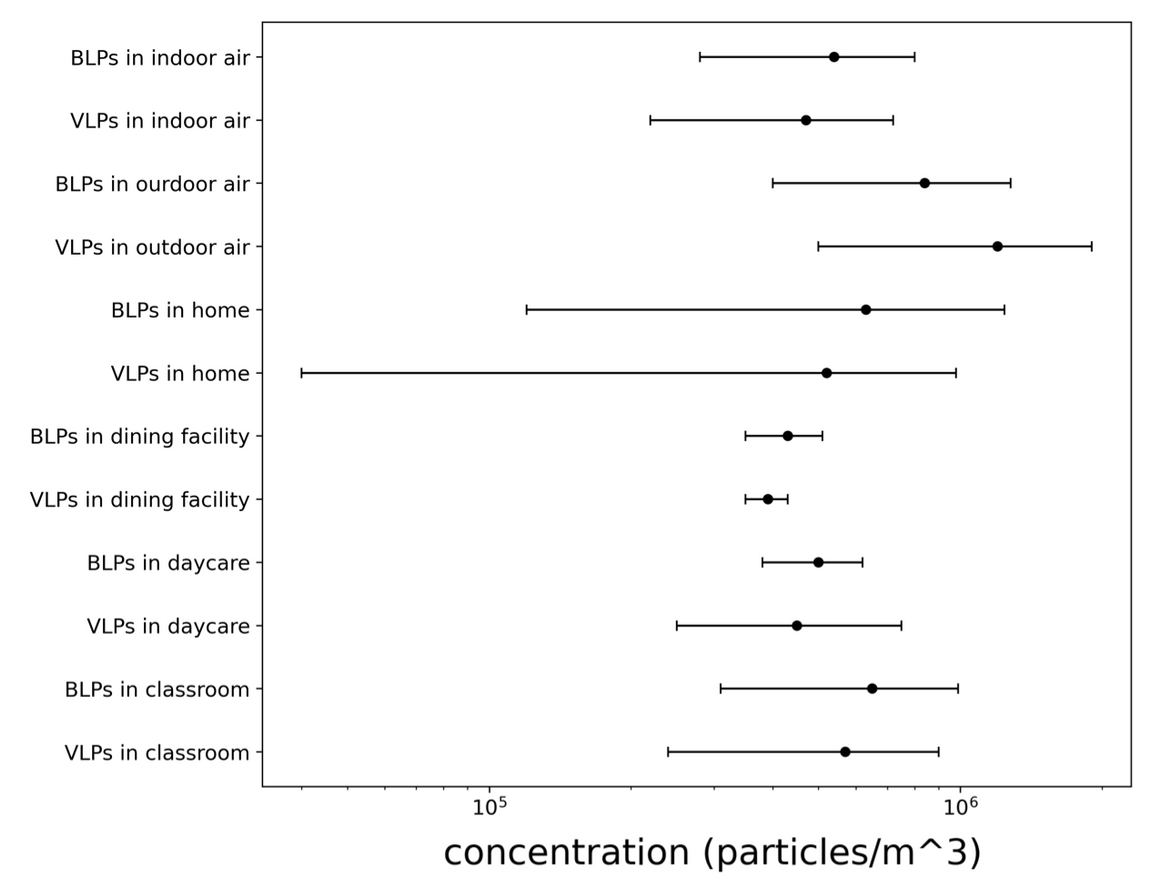
Figure A2-1: Concentration of airborne virus- and bacteria-like particles in different locations measured by Prussin et al. 2015. Microbes were counted using the SYBR Gold fluorescence dye to stain nucleic acid. Fluorescent particles between 0.02 and 0.50 μm were counted as VLPs and those between 0.50 and 5.00 μm as BLPs. Error bars represent the max-min concentrations measured in samples from the same location.
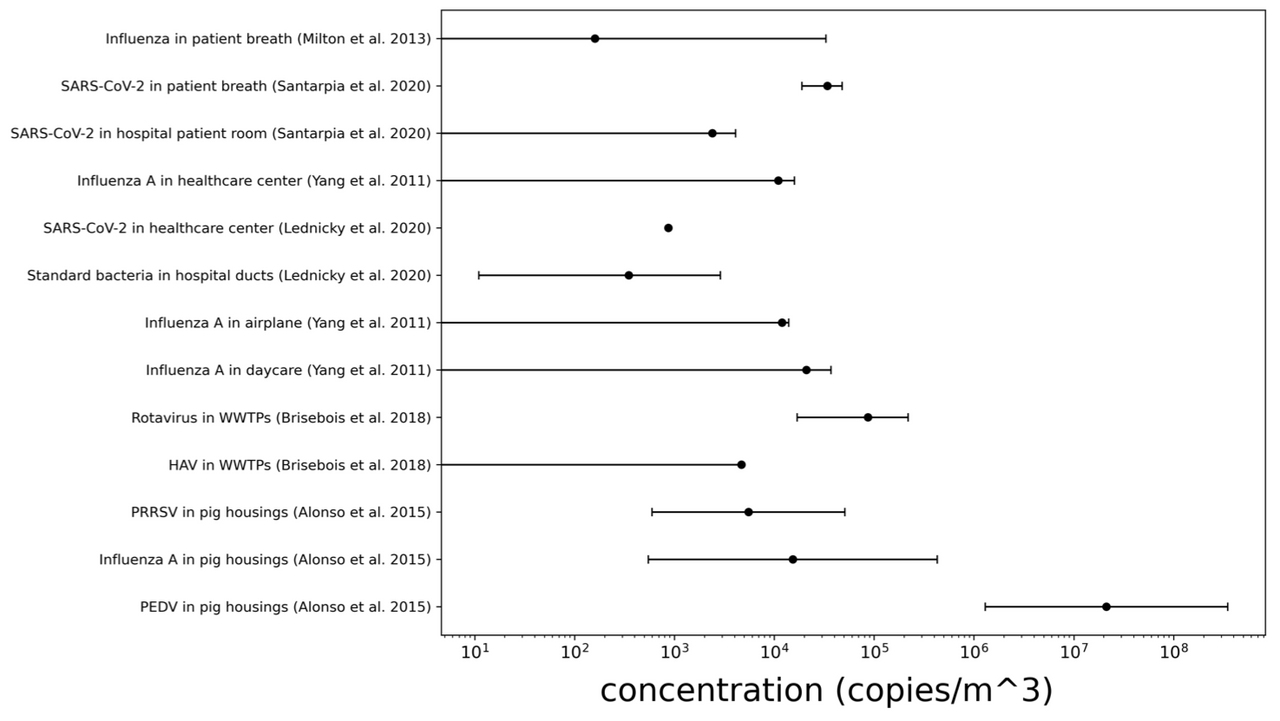
Figure A2-2: Concentration of airborne pathogens by qPCR. Error bars represent the max-min concentrations measured in samples from the same study. A concentration of 0 means that a “no detect” (nd) was also measured in this location.
Appendix 3: Outdoor air sampling
Samples could be gathered outdoors in major cities and travel hubs (10, 76, 78–87). Sampling in these urban areas offers a wide geographic reach and a large population catchment. There is a possibility that if a substantial number of people in a city were infected and shedding pathogens, strategically placed high-volume air samplers might detect bioaerosols in the outdoor environment. However, the likelihood of encountering human-relevant pathogens in outdoor air is generally lower, leading to my skepticism about the effectiveness of this approach.
Moreover, outdoor sampling is particularly susceptible to environmental variables, more so than other locations. When conducting time-series detection approaches with metagenomic air sampling, it’s crucial to account for environmental factors that could skew the abundance of the target pathogen. Outdoors, the presence of numerous potential confounding factors complicates this process.
Despite these challenges, various government-funded biosurveillance initiatives are focused on outdoor air sampling. It remains unclear whether these programs are specifically aimed at detecting aerial bioweapon attacks, such as anthrax-laden warheads, or if there is an unrecognized potential in outdoor air sampling that surpasses current evidence.
Notable programs:
- The SIGMA+ program from DARPA employs outdoor air collection through emergency vehicles equipped with custom air samplers and portable samplers like the pBDi.
- The U.S. government's BioWatch program initially utilized existing EPA air sampling sites and collectors, predominantly outdoor. Many of these were located near airports and other urban centers (6, 88, 89). The current sampling locations of BioWatch are not publicly available.
- The Kromek Biosequencer is an innovative system, attaching an air sampler to a vehicle, integrating automated sample preparation and long-read sequencing capabilities..
Appendix 4: In-duct sampling
Heating, ventilation, and air conditioning (HVAC) systems present a viable option for filter-based bioaerosol collection due to several factors: 1) the prevalence of existing HVAC systems with filters in many buildings, allowing for non-invasive sampling; 2) their capacity to handle large air volumes; and 3) their ability to collect particles from extensive spatial areas within buildings. Indeed, numerous studies have investigated microbial communities in HVAC systems, either by attaching external air sampling devices to these systems (16, 90) or by sampling directly from HVAC filters (5, 77, 91–96). Some of these studies, exploring viral diversity, have successfully identified various human pathogens, including influenza, SARS-CoV-2, retroviruses, adenovirus, and RSV, in HVAC systems (5, 16, 77, 96).
However, there are uncertainties regarding the practicality of using HVAC filters for bioaerosol sampling. Firstly, viral concentrations in HVAC filters are generally low, a characteristic common to many air samples. Additionally, there is some indication, albeit not strong, that nucleic acid (NA) concentrations within ducts may be lower than those found in-room (74, 93). Secondly, most studies on HVAC filters span several months, aligning with the typical frequency of filter replacement in these systems (every three months). This raises questions about the ability of HVAC filters to collect adequate NA over shorter time frames. While using higher efficiency filters, like MERV 12 or above, might enhance virus collection, this often isn't cost-effective in larger buildings due to increased power requirements (4).
In conclusion, I anticipate that in-room sampling methods will prove more promising than HVAC sampling. This is because in-room methods offer greater flexibility in adjusting sampling device properties, such as flow rate, and allow for more strategic selection of sampling locations.
Appendix 5: NA recovery BOTEC
Concentrations
-
BLPs in indoor environment = 540 BLP/L (Prussin et al. 2015)
-
VLPs in indoor environment = 470 VLP/L (Prussin et al. 2015)
Genome
-
Typical bacterial genome = 5E6 bp/BLP
-
Typical viral genome = 4E4 bp/VLP
-
Mass of single bp = 1.08E-12 ng/bp
Air sampling
-
Flow rate of air sampler = 200 L/min = 3.33 L/s (ACD 200 Bobcat air sampler)
-
Sampling time = 8 hr = 28800 s
NA recovery
NA recovery rate = Flow rate × Microbe concentration × Typical genome mass
NA recovery rate = (3.33 L/s)×[(5E6 bp/BLP)×(540 BLP/L) + (4E4 bp/VLP)×(470 VLP/L)]×(1.08E-12 ng/bp)
NA recovery rate = 0.0098 ng/s
NA recovered over 8 hours = 280 ng
Assumptions
-
Each VLP and BLP contains only one standard viral or bacterial genome. In reality, airborne particles often contain multiple microbes. This assumption pushes the results in the “more NA” direction.
-
Microbe concentrations remain unchanged as a result of air sampling, which is probably a valid assumption in large indoor spaces, but doesn’t hold in more confined environments. This assumption pushes the results in the “less NA” direction.
Footnotes
-
↩
Passive air sampling is a branch of air sampling that depends on natural settling of particles. Methods for passive sampling include surface dust collection (e.g., vacuuming), settle plates, and adhesive tape.
-
↩
In the case of filter-based sampling, for example, higher flow rates trade off against increased destructiveness, as the high air flow rate causes mechanical stress that kills microbes and damages nucleic acid. Additionally, filters-based methods can achieve high capture efficiency at the small particle sizes characteristic of viral aerosols, but this introduces limits on the flow rate. Similar tradeoffs exist for other sampler categories (see Appendix 2).
-
↩
For example, an electrostatic precipitator might deposit particles into a liquid medium, or a condensation sampler might have an initial filter to remove larger particles.
-
↩
Microbes were counted using the SYBR Gold fluorescence dye to stain nucleic acid. Fluorescent particles between 0.02 and 0.50 μm were counted as VCPs and those between 0.50 and 5.00 μm as BCPs.
-
↩
A simple BOTEC suggests that eight hours of air sampling in an indoor environment with typical bacteria and virus concentrations at 200 L/min would yield about 300 ng of NA (Appendix 5). It’s not uncommon to see amplification-free metagenomic protocols call for at least 1 μg (or 1000 ng) of NA.
-
↩
It generally requires more power to drive air through filters with small pore diameters.
-
↩
In reality, “inertia” is too simple a concept to characterize the movement of particles along streamlines. A particle’s Stokes number provides a more rigorous explanation of its behavior (56).
-
↩
A large pressure drop like that caused by a filter with a small effective pore size requires more power to push air through the pressure interface.